Neural Drug Discovery
July 2021
De novo drug discovery is traditionally a computationally expensive, slow, and costly, process. At a high level, it can be thought of as creating some drug that binds to a given protein. Recently, there have been large steps taken forward in both in silico drug discovery and neural machine translation (NMT). So combine the two. In this project, I formulate generating a drug given a protein as a translation task, and apply recent NMT methods to approach this problem. With some domain-specific modifications, this model performs well, and may be used as a foundation of de novo drug discovery techniques. Testing on a curated set of proteins with known binders, the approach was able to generate molecules have good binding energies and attractive computed attributes, such as synthetic accessibility scores and drug likeness (QED).
Introduction
-
Drug development is expensive
-
There is a lot to go through (chemical space). Beyond that, the space of chemicals of interests is incredibly sparse. There’s an estimated \(10^{60}\) molecules under 500 molecular weight, yet an extremely small fraction of those are druglike.
-
We need better ways to explore such chemical space effectively and cost-efficiently.
-
So let’s pretend our protein : ligand pairs are sentences in different languages, and translate between the two using recent NLP techniques.
Methodology
Put a little more formally, we have pairs of known proteins and ligands, with the corresponding interaction strength. Given some protein, we wish to generate some ligand which binds to it. Fortunately, we can represent both proteins and ligands in a canonical text format. So let’s pretend we’re simply translating between the two “languages”.
Dataset
To get good protein : ligand pairs, the BindingDB database1 was used. The BindingDB database is a collection of ~800k protein : ligand pairs, with associated recorded binding strengths. However, many of these (the majority) bind very weakly, and much of the data is redundant. So let’s preprocess.
-
Canonicalize all proteins and ligands;
-
Remove all amino acids and ligands with length greater than 1024 characters;
-
Remove all invalid ligands and proteins;
-
Remove all duplicate protein-ligand pairs;
-
Remove all protein-ligand pairs with \(K_\text{i}\), \(K_\text{d}\), IC\(_{50}\), or EC\(_{50}\) values above 100 nM;
-
Use the BLASTP algorithm to remove all proteins that had greater than 90% identity with our testing set.
The testing dataset was composed of 62 unique proteins and 186 ligands from the “core” partition of the PDBbind dataset2. The “core” partition itself is a high-quality set of highly diverse proteins, created primarily for the task of benchmarking docking/scoring methods. We instead use it as a reference to compare against for the generated molecules. Since BindingDB is a superset of PDBBind, step (6) above removes any possible data leaks from the training set to the test set.
Model hyperparameters
For the input amino acids, we treated each character as a token. For the
output SMILES, we replaced functional group substrings with special
tokens, replacing the largest to the smallest, then treated each
non-special character as a unique token. We used global-chem for the
list of functional groups.
In the base model, a transformer3, we used an embedding dimension of 128, feed-forward dimension of 128, 8 encoder and decoder layers with 4 attention heads per attention block. Additionally, we used dropout of 0.1, AdamW optimizer with betas 0.9 and 0.98, a weight decay of 0.01, base learning-rate of 0.0005 and an inverse square-root warm-up. Batch-size was set to 40,000 tokens. For decoding, we use top-p (nucleus) sampling4 with \(p = .95\). We generated 40 different molecules per protein during evaluation, although this could easily be extended to thousands or beyond, with performance of the model only increasing as number of molecules returned does. To implement the model, we used fairseq5. All model training was done in under 8 hours on a single V100 GPU.
Molecule generation
We found that, when using beam search, the transformer decoder commonly degenerated into repeating sequences, commonly carbons; switching to top-p fixed this.
After generation, we filter the output using traditional high throughput screening (HTS) methods with RDKit:
-
We remove all invalid molecules, and molecules with synthetic accessibility (SA) scores above 5,
-
We filter remaining molecules for PAINS and BRENK substructures,
-
We then sort by QED and select the top \(n\) molecules with the highest QED for each protein. We use \(n = 10\) due to computational restraints.
Molecule scoring
For the protein-ligand docking simulation, we use CB-Dock6. F or the input, we use the protein pocket given by PDBBind and specify we only want a single cavity; CB-Dock automatically generates the bounding box. We bind each protein against all ligands generated targeted at that protein, in addition to the ground truth known ones for reference.
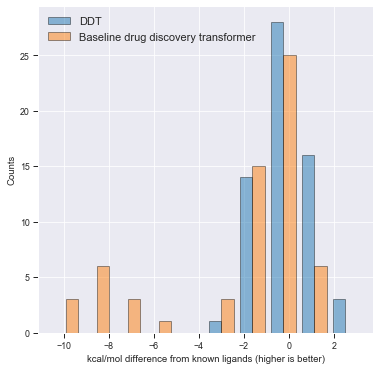
In the above figure, the “baseline drug discovery transformer” is a naive character-level implementation of the above process using beam-search instead of top-p. The baseline binding strength for each protein is the strongest binding ligand given by PDBBind for that given pocket. We subtract the score of the strongest binder from the binding energy of generated molecules for fair comparison.
Conclusion
Applying NMT techniques works even to far out-of-domain problems. This project explored domain specific engineering and preparation of data, the generalization of NMT techniques to biology, and the general processing of going from an idea to presentable results, including data gathering, creating an evaluation metric, and producing measureable and significant results.